Large dataset on 8GB RAM? Let IterableDataset handle
Context
I am currently working on a directed-message passing neural network called Chemprop, which is a useful model for structure-property prediction. As a student, my resources are quite limited, with only 8GB or 16GB of RAM available to me. However, I need to handle a dataset containing 1 million records for my project.
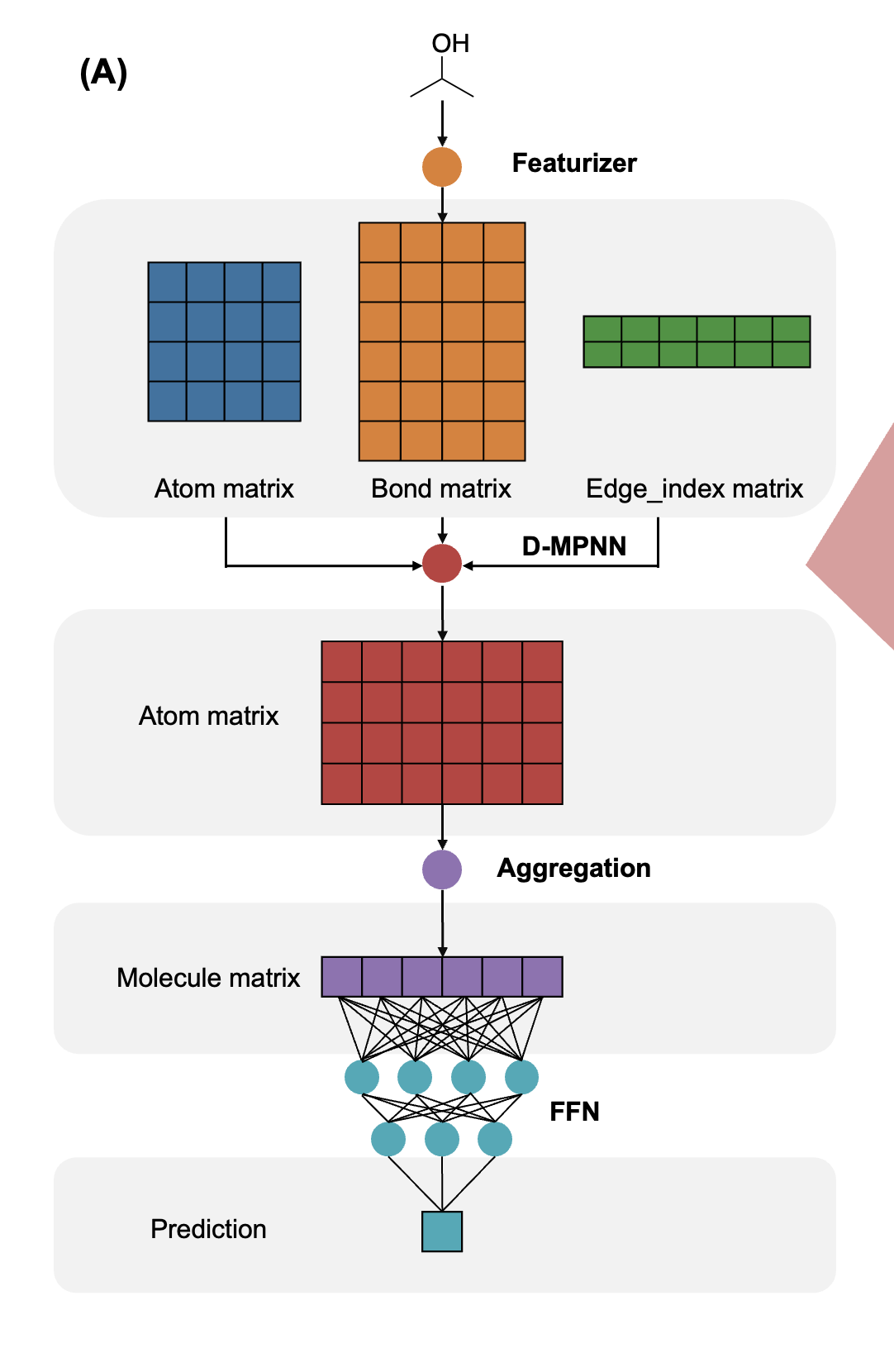
To use Chemprop, the input consists of SMILES strings and target values stored in a CSV file. These SMILES strings are then converted into a MoleculeDataset, which includes information about the molecular graph, target values, and more. The MoleculeDataset is subsequently passed to a DataLoader to create batches for the training loops.
With my 8 GB RAM laptop, I can load the entire CSV file, but converting all the data into a MoleculeDataset at once is not feasible. Therefore, I needed to find an alternative approach to:
- Handle this conversion step
- Compatible with Chemprop framework.
Eventually, I discovered the IterableDataset feature in PyTorch, which effectively resolves this issue. All the scripts below can be found on my GitHub.
Scripts
First, I need to define some useful functions to transform a Pandas DataFrame containing SMILES strings into a MoleculeDataset. If we simply call the `dataset_preparator()` function, the entire Pandas DataFrame will be converted into a MoleculeDataset, which is not feasible given my 8GB of RAM.
import pandas as pd
from chemprop import data, featurizers
import pandas as pd
import torch
from torch.utils.data import IterableDataset
from chemprop import data, featurizers
from sklearn.preprocessing import StandardScaler
import psutil
import os
import gc
import time
import numpy as np
def datapoint_preparator(df,smiles_column,target_column):
smis = df.loc[:,smiles_column].values
ys = df.loc[:,[target_column]].values
datapoints = [data.MoleculeDatapoint.from_smi(smi,y) for smi, y in zip(smis,ys)]
return datapoints
def dataset_preparator(df, smiles_column, target_column, featurizer = featurizers.SimpleMoleculeMolGraphFeaturizer()):
datapoints = datapoint_preparator(df=df, smiles_column=smiles_column, target_column=target_column)
dataset = data.MoleculeDataset(datapoints, featurizer=featurizer)
return datasetThus, the strategy utilizing PyTorch’s IterableDataset has been established.
class IterableMolDatapoints(IterableDataset):
'''A class to prepare data for streaming, which is a subclass of IterableDataset.
The output is a generator that yields one chemprop.data.datasets.Datum at a time.
'''
def __init__(self, df, smiles_column, target_column, scaler = None, size_at_time=100, shuffle=True):
'''Parameters:
----------
df (pd.DataFrame): A pandas dataframe containing the data.
smiles_column (str): The column name containing SMILES strings.
target_column (str): The column name containing the target values.
scaler (StandardScaler): A StandardScaler object (already fitted) for normalizing the target values.
size_at_time (int): The number of samples to transfrom into chemprop.data.datasets.Datum at a time.
shuffle (boolean): If the df is shuffled.'''
super().__init__()
self.df = df
self.smiles_column = smiles_column
self.target_column = target_column
self.size_at_time = size_at_time
self.shuffle= shuffle
self.scaler = scaler
def __len__(self):
return len(self.df)
def __iter__(self):
'''A function to define iteration logic. It take the whole csv data, then shuffled, then access to only a subset of data at a time for transformation.
The output is a generator that yields chemprop.data.datasets.Datum and ready to put through DataLoader.
'''
if self.shuffle:
df_shuffled = self.df.sample(frac=1).reset_index(drop=True)
else:
df_shuffled = self.df.copy()
# Transform pandas dataframe to molecule dataset according to size_at_time, prevent overloading memory. This is to balance between memory and speed.
for i in range(0, len(df_shuffled), self.size_at_time):
df_at_time = df_shuffled.iloc[i:i + self.size_at_time]
df_process = dataset_preparator(df=df_at_time, smiles_column=self.smiles_column, target_column=self.target_column)
if self.scaler != None:
df_process.normalize_targets(self.scaler)
# Handling parallelization manually
worker_info = torch.utils.data.get_worker_info()
if worker_info is None:
for mol in df_process:
yield mol
else:
num_workers = worker_info.num_workers
worker_id = worker_info.id
for i, mol in enumerate(df_process):
if i % num_workers == worker_id:
yield molHere is an example of how to use the IterableMolDatapoints class. Please note that if you want to normalize the target values, you must do it manually and then pass the scaler object to IterableMolDatapoints.
# Prepare Data
smiles_column = 'smiles'
target_column = 'docking_score'
df_train = pd.read_csv('on_the_fly_data.csv')
scaler = StandardScaler().fit(df[[target_column]])
# Create iterable data
iterable_dataset = IterableMolDatapoints(
df=df_train,
smiles_column=smiles_column,
target_column=target_column,
size_at_time=100, scaler=scaler, shuffle=True
)
iterable_train_loader = data.build_dataloader(
iterable_dataset,
batch_size=5, shuffle=False)
# Doing something with DataLoader
for epoch in range(50):
for batch in iterable_train_loader:
# Train model or doing something hereAnother important point to mention is the `size_at_time` parameter. This parameter helps balance memory usage with computation rate. If we set `size_at_time` to 1, the class behaves like a default `IterableDataset`, processing one record at a time. However, if we increase `size_at_time` to a higher value, say 100, the transformation process will operate on 100 samples simultaneously, holding them until the DataLoader calls them out. While this approach requires more memory, it can lead to faster DataLoader calls.
The usage of an `IterableDataset` is similar to that of a `MapStyle Dataset` (the default indexable dataset), as illustrated below.
# Prepare Data
smiles_column = 'smiles'
target_column = 'docking_score'
df_train = pd.read_csv('on_the_fly_data.csv')
scaler = StandardScaler().fit(df[[target_column]])
# Create map data
map_dataset = dataset_preparator(df_train, smiles_column, target_column)
map_dataset.normalize_targets(scaler)
map_loader = data.build_dataloader(map_dataset, batch_size=5, shuffle=False)
# Doing something with DataLoader
for epoch in range(50):
for batch in map_loader:
# Train model or doing something here
Two main challenges: Shuffling and Parallelization
There are two main challenges with the iterable dataset: shuffling and parallelization. Since I can load the entire Pandas dataframe, I manually shuffle it using this line.
df_shuffled = self.df.sample(frac=1).reset_index(drop=True)This can be specified by the shuffle argument when initializing the class. It is important to note that shuffling will be managed within IterableDataset; when using DataLoader, the shuffle argument should always be disabled.
When parallelizing, IterableDataset has significant issues with num_workers > 0. Because IterableDataset cannot be accessed by index, it cannot sample indices and distribute them to each worker. Therefore, we need to handle this manually in the __iter__() method.
worker_info = torch.utils.data.get_worker_info()
if worker_info is None:
for mol in df_process:
yield mol
else:
num_workers = worker_info.num_workers
worker_id = worker_info.id
for i, mol in enumerate(df_process):
if i % num_workers == worker_id:
yield molPerformance test
I have conducted three tests regarding the IterableDataset implementation.
Test 1: Memory usage when creating Datasets
# Prepare data
data_path = 'on_the_fly_data.csv'
smiles_column = 'smiles'
target_column = 'docking_score'
df = pd.read_csv(data_path)
df = df.sample(100000)
scaler = StandardScaler().fit(df[[target_column]])
# Function to record memory
def memory_record():
process = psutil.Process(os.getpid())
mem = process.memory_info().rss / 1024 ** 2 # in MB
return mem
# For Iterable Dataset
gc.collect()
start_time = time.time()
memory_before = memory_record()
iterable_dataset = IterableMolDatapoints(
df=df,
smiles_column=smiles_column,
target_column=target_column,
size_at_time=100, scaler=None, shuffle=True
)
memory_after =memory_record()
end_time = time.time()
gc.collect()
print(f'Memory usage to load iterable dataset: {memory_after-memory_before} MB ')
print(f'Time to load iterable dataset: {end_time-start_time} s ')
# For Mapstyle Dataset
gc.collect()
start_time = time.time()
memory_before = memory_record()
dataset = dataset_preparator(
df=df,
smiles_column=smiles_column,
target_column=target_column
)
memory_after = memory_record()
end_time = time.time()
gc.collect()
print(f'Memory usage to load map dataset: {memory_after-memory_before} MB ')
print(f'Time to load map dataset: {end_time-start_time} s ')Memory usage to load iterable dataset: 0.0 MB
Time to load iterable dataset: 0.0011718273162841797 s
Memory usage to load map dataset: 677.5 MB
Time to load map dataset: 9.14915418624878 s
Test 2: Similar Behavior when passing IterableDataset and Mapstyle Dataset to DataLoader
# Prepare Data
smiles_column = 'smiles'
target_column = 'docking_score'
df_train = pd.read_csv('on_the_fly_data.csv')
df_train = df_train.sample(1000) # Sample a small subset for illustration
# Create map data
map_dataset = dataset_preparator(df_train, smiles_column, target_column)
map_loader = data.build_dataloader(map_dataset, batch_size=5, shuffle=False)
# Create iterable data
iterable_dataset = IterableMolDatapoints(
df=df_train,
smiles_column=smiles_column,
target_column=target_column,
size_at_time=5, shuffle=False, #scaler=scaler
)
iterable_loader = data.build_dataloader(iterable_dataset, batch_size=5, shuffle=False)
def compare_loader(loader1, loader2):
"""
Check if two data loaders produce the same data in the same order.
Parameters:
- loader1, loader2: DataLoader instances to compare
Returns:
- bool: True if loaders produce identical data
"""
# If they have the same lengh:
if len(loader1) != len(loader2):
print(f"Loaders have different lengths: {len(loader1)} vs {len(loader2)}")
return False
# Compare each batch attribute
for i, (batch1, batch2) in enumerate(zip(loader1, loader2)):
# Compare MolGraph objects
same_nodes = np.array_equal(batch1.bmg.V, batch2.bmg.V)
same_edges = np.array_equal(batch1.bmg.E, batch2.bmg.E)
if same_nodes and same_edges:
print(f"MolGraphs are identical in batch {i}")
else:
print(f"MolGraphs are different in batch {i}")
return False
# Compare targets
same_target = np.array_equal(batch1.Y, batch2.Y)
if same_target:
print(f"Targets are identical in batch {i}")
else:
print(f"Targets are different in batch {i}")
return False
# Compare more attributes if needed
return True
# Test the similarity between the two data loaders:
iterable_loader = data.build_dataloader(iterable_dataset, batch_size=2, shuffle=False)
map_loader = data.build_dataloader(map_dataset, batch_size=2, shuffle=False)
if compare_loader(iterable_loader, map_loader):
print("The data loaders contain the same data in the same order")
else:
print("The data loaders differ")MolGraphs are identical in batch 0
Targets are identical in batch 0
MolGraphs are identical in batch 1
Targets are identical in batch 1
...
MolGraphs are identical in batch 499
Targets are identical in batch 499
The data loaders contain the same data in the same order
Test 3: Shuffle ability
# Prepare Data
smiles_column = 'smiles'
target_column = 'docking_score'
df_train = pd.read_csv('on_the_fly_data.csv')
df_train_10 = df_train.sample(10) # Take a small subset for example
# Create iterable data
iterable_dataset = IterableMolDatapoints(
df=df_train_10,
smiles_column=smiles_column,
target_column=target_column,
size_at_time=5, scaler=None, shuffle=True
)
iterable_train_loader = data.build_dataloader(
iterable_dataset,
batch_size=5, shuffle=False)
print('Data batches with Unscaled target values:')
for epoch in range(2):
print(f'Epoch {epoch+1}')
for i, batch in enumerate(iterable_train_loader):
print(f'Batch {i+1}')
print(batch.Y)Data batches with Unscaled target values:
Epoch 1
Batch 1
tensor([[-6.2346],
[-5.8180],
[-7.0793],
[-7.8406],
[-6.3524]])
Batch 2
tensor([[-6.5732],
[-5.1087],
[-6.5070],
[-4.9264],
[-7.2945]])
----------------------------------------
Epoch 2
Batch 1
tensor([[-7.0793],
[-6.5070],
[-6.3524],
[-6.2346],
[-5.8180]])
Batch 2
tensor([[-4.9264],
[-7.2945],
[-7.8406],
[-5.1087],
[-6.5732]])
----------------------------------------
Key messages
IterableDataset helps prevent data overloading. We can choose how many data samples to process at a time by specifying size_at_time.
Shuffle and parallelization with IterableDataset is challenging due to no indexability.
Final thoughts
I understand that every approach has its pros and cons, and programming is about generalization and optimization. That’s why I want to bring this idea to the discussion. What do you think?
Here are some limitations of the current version of my code that I’ve identified:
It works well for molecular properties prediction tasks; however, for multicomponent tasks, some adaptation may be needed.
It still requires access to the entire CSV file at the beginning to shuffle the data.
Comments
Post a Comment