Integrate cheminformatics data and ClaudeAI— Part 1: ChEMBL
First appearing in November 2024, Model Context Protocol (MCP) quickly gained popularity within the AI community. This innovative technology allows for the integration of various resources, such as databases, local documents, and websites, into LLMs. As a result, AI applications can access more up-to-date data that might have previously been restricted or unseen.
In this post, I will share my experience integrating ClaudeAI with three of the most popular cheminformatics databases: ChEMBL, PubChem, and Protein Data Bank (PDB). Concomitantly, I also share my view about the pros and cons of MCP. All the code can be found on my GitHub. I hope you find this instruction helpful!
Fundamental Concepts
Model Context Protocol (MCP) is a protocol that allows large language models (LLMs) to connect to external tools, APIs, or environments to access resources, task-specific context, and perform actions outside of their native model boundaries. It essentially augments the model’s capabilities by adding context improves the model’s accuracy, reasoning, and usefulness in complex tasks.
This is a very brief description of MCP. To keep this blog from being too long, I suggest you read more about MCP on their original website.
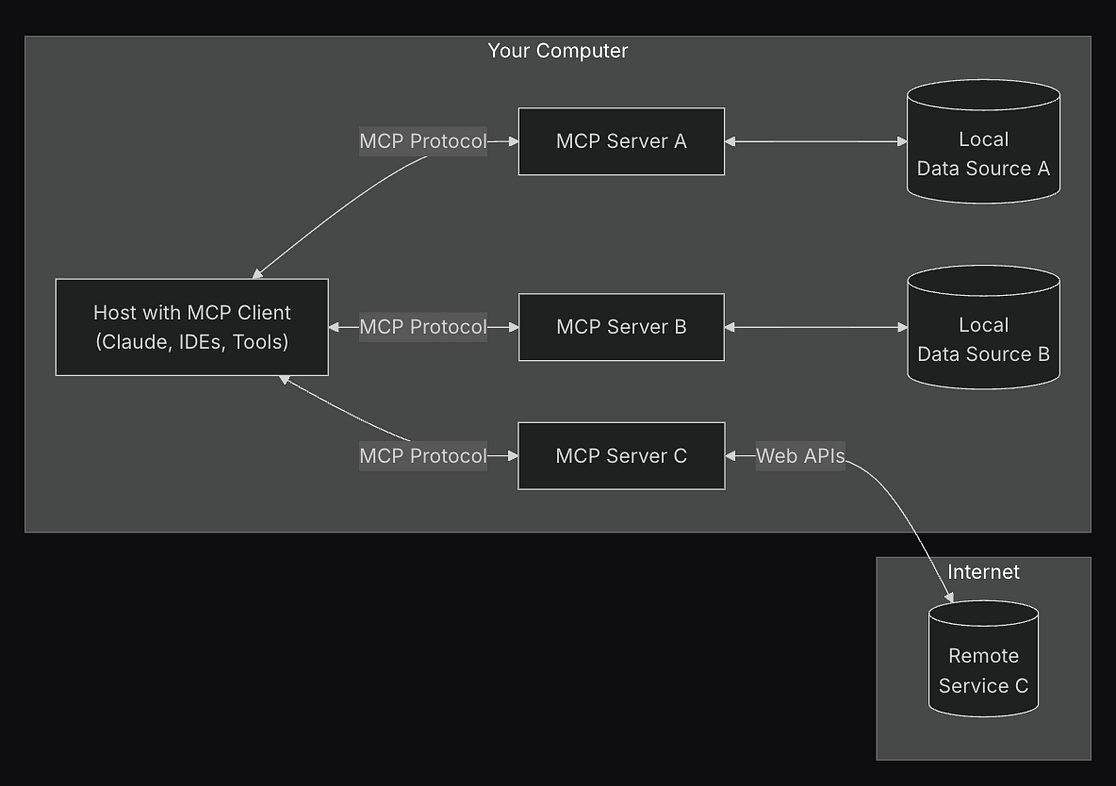
ChEMBL Data Sources: Web service or Database?
There are two ways to integrate ChEMBL data into the MCP server:
Using the Web Service: The main advantage of using the web service is that the data is always up-to-date and directly compatible with the latest version of ChEMBL, which means that there is no need for manual updates. However, there are several limitations to consider. The ChEMBL API enforces a rate limit of 1 request per second for users without an API key, which can significantly slow down large-scale automated data retrieval.
Using the ChEMBL Database: In this approach, MCP is connected directly to a locally hosted ChEMBL database. The major benefit is unrestricted access to the entire dataset at once, which allows for high-throughput queries, large-scale searches, and complex manipulations without API rate limitations. However, this comes with the trade-off of manually downloading, hosting, and updating the ChEMBL database every time a new version is released, which requires additional setup and maintenance effort.
Although I really want to use the ChEMBL Database approach, it is impossible for me due to my storage limit. The dump file of ChEMBL database costs 17.5GB, which might expand to 70+ GB for the full database. Additionally, I also want to integrate PubChem and PDB. Therefore, using an API call through a web service is my only choice.
Note: If you are interested in full database implementation and you have sufficient resources, we can contact haha.
Integrating MCP and CHEMBL Web Services
I also followed the basic MCP instruction for Python scripts. If you simply want to create an MCP server that connects to your local folder, the task can be much simpler.
1. Install Claude Desktop, uv, and dependencies:
First, we need to install Claude Desktop on our computer. You can find the download instructions here. After having Claude Desktop, you need to install uv:
curl -LsSf https://astral.sh/uv/install.sh | shThen, we need to initialize the directory for our project, let's call it cheminformatics_mcp:
# Create a directory for project
uv init cheminformatics_mcp
cd cheminformatics_mcp
# Create virtual environment and activate it
uv venv
source .venv/bin/activate
# Install dependencies, you can try more than that
uv add "mcp[cli]" httpx
# Create our server file
touch cheminformatics_mcp.py2. Establish the MCP server script:
This is the fundamental code in the cheminformatics_mcp.py script:
from typing import Any
import httpx
import asyncio
from urllib.parse import quote
from mcp.server.fastmcp import FastMCP
mcp = FastMCP('cheminformatics_mcp')
# Install some tools/resources/prompt here
# ....
if __name__ == "__main__":
mcp.run(transport='stdio')3. Establish Tools:
The tools are functions that can be called by the LLM with user approval. It is similar to the way we use Python scripts to access the ChEMBL web service. The only difference is that we add @mcp.tool() decorators for the scripts.
Let's have a look at the example below. I have a script to fetch data given any CHEMBL_ID. This is just a Python script, nothing to do with MCP. The script will use chembl_id_lookup to determine which resources the CHEMBL_ID belongs to (such as molecule, target, or assay), then use that resource to access the data.
async def fetch_chembl_id(chembl_id: str, timeout: float = 10.0):
base_url = "https://www.ebi.ac.uk"
lookup_url = f"{base_url}/chembl/api/data/chembl_id_lookup/{chembl_id}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
# Lookup the object type of CHEMBLID
lookup_response = await client.get(lookup_url)
lookup_response.raise_for_status()
lookup_data = lookup_response.json()
# Establish object url base on lookup result
resource_url = lookup_data.get("resource_url") + ".json"
if not resource_url:
print(f"No resource_url found for {chembl_id}")
return None
full_url = base_url + resource_url
resource_response = await client.get(full_url)
resource_response.raise_for_status()
return resource_response.json()
except httpx.HTTPStatusError as e:
print(f"HTTP error {e.response.status_code} for {chembl_id}: {e.response.text}")
except Exception as e:
print(f"Unexpected error for {chembl_id}: {e}")
return NoneTo make this become to be able for the MCP server to access, we add it to the cheminformatics_mcp.py with @mcp.tool() decorator:
from typing import Any
import httpx
import asyncio
from urllib.parse import quote
from mcp.server.fastmcp import FastMCP
mcp = FastMCP('cheminformatics_mcp')
# Install some tools/resources/prompt here
@mcp.tool()
async def fetch_chembl_id(chembl_id: str, timeout: float = 10.0):
base_url = "https://www.ebi.ac.uk"
lookup_url = f"{base_url}/chembl/api/data/chembl_id_lookup/{chembl_id}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
# Lookup the object type of CHEMBLID
lookup_response = await client.get(lookup_url)
lookup_response.raise_for_status()
lookup_data = lookup_response.json()
# Establish object url base on lookup result
resource_url = lookup_data.get("resource_url") + ".json"
if not resource_url:
print(f"No resource_url found for {chembl_id}")
return None
full_url = base_url + resource_url
resource_response = await client.get(full_url)
resource_response.raise_for_status()
return resource_response.json()
except httpx.HTTPStatusError as e:
print(f"HTTP error {e.response.status_code} for {chembl_id}: {e.response.text}")
except Exception as e:
print(f"Unexpected error for {chembl_id}: {e}")
return None
if __name__ == "__main__":
mcp.run(transport='stdio')Following this step, we can implement whatever tools we like. For developers, you can have a look at the ChEMBL Web Service to establish more tools. This is my full script with 4 basic tools:
import httpx
from mcp.server.fastmcp import FastMCP
import asyncio
from urllib.parse import quote
mcp = FastMCP('cheminformatics_mcp')
valid_resources = [
"activity", "assay", "atc_class", "binding_site", "biotherapeutic", "cell_line",
"chembl_id_lookup", "compound_record", "compound_structural_alert", "document",
"document_similarity", "document_term", "drug", "drug_indication", "drug_warning",
"go_slim", "mechanism", "metabolism", "molecule", "molecule_form", "organism",
"protein_classification", "similarity", "source", "status", "substructure", "target",
"target_component", "target_relation", "tissue", "xref_source"
]
# Tool 1
@mcp.tool()
async def fetch_chembl_id(chembl_id: str, timeout: float = 10.0):
"""
Fetches detailed information for a ChEMBL ID by first looking up the ID's resource type,
then retrieving the full data from the appropriate endpoint.
Parameters:
-----------
chembl_id : str
A valid ChEMBL identifier
timeout : float, optional
Maximum time in seconds to wait for the API responses (default: 10.0)
Returns:
--------
Optional[Dict[str, Any]]
The complete data for the ChEMBL ID as a dictionary if successful, None otherwise
"""
base_url = "https://www.ebi.ac.uk"
lookup_url = f"{base_url}/chembl/api/data/chembl_id_lookup/{chembl_id}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
lookup_response = await client.get(lookup_url)
lookup_response.raise_for_status()
lookup_data = lookup_response.json()
resource_url = lookup_data.get("resource_url") + ".json"
if not resource_url:
print(f"No resource_url found for {chembl_id}")
return None
full_url = base_url + resource_url
resource_response = await client.get(full_url)
resource_response.raise_for_status()
return resource_response.json()
except httpx.HTTPStatusError as e:
print(f"HTTP error {e.response.status_code} for {chembl_id}: {e.response.text}")
except Exception as e:
print(f"Unexpected error for {chembl_id}: {e}")
return None
# Tool 2
@mcp.tool()
async def fetch_chembl_given_resources(chembl_id: str, resources: str, timeout: float = 10.0):
"""
Fetches data directly from a specific ChEMBL API resource endpoint for a given ChEMBL ID.
Use this function when you already know which resource type the ChEMBL ID belongs to.
Parameters:
-----------
chembl_id : str
A valid ChEMBL identifier
resources : str
The specific ChEMBL resource type to query. Must be one of the valid resources listed
in the valid_resources list (e.g., 'molecule', 'target', 'assay')
timeout : float, optional
Maximum time in seconds to wait for the API response (default: 10.0)
Returns:
--------
Optional[Dict[str, Any]]
The data from the specified resource for the ChEMBL ID as a dictionary if successful,
None otherwise
"""
if resources not in valid_resources:
print(f"Invalid resource: {resources}. Valid resources are: {valid_resources}")
return None
url = f"https://www.ebi.ac.uk/chembl/api/data/{resources}/{chembl_id}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
response = await client.get(url)
response.raise_for_status()
return response.json()
except httpx.HTTPStatusError as e:
print(f"HTTP error {e.response.status_code} for {chembl_id}: {e.response.text}")
except Exception as e:
print(f"Unexpected error for {chembl_id}: {e}")
return None
# Tool 3
@mcp.tool()
async def substructure_search(molecule: str, limit: int = 10, timeout: float = 10.0, offset: int = 0):
"""
Performs a substructure search in the ChEMBL database given a molecule.
This function finds molecules in the ChEMBL database that contain the specified
substructure pattern defined by the SMILES string.
Parameters:
-----------
molecule : str
The SMILES, CHEMBL_ID or InChIKey
limit : int, optional
Maximum number of results to return (default: 10)
timeout : float, optional
Maximum time in seconds to wait for the API response (default: 10.0)
offset : int, optional
Number of results to skip for pagination (default: 0)
Returns:
--------
Optional[List[Dict[str, str]]]
A list of dictionaries containing the ChEMBL ID and canonical SMILES
for each matching molecule if successful, None otherwise.
Each dictionary has keys 'CHEMBL_ID' and 'SMILES'.
"""
encoded_molecule = quote(molecule)
url = f"https://www.ebi.ac.uk/chembl/api/data/substructure/{encoded_molecule}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
response = await client.get(url, params={"limit": limit, "offset": offset})
response.raise_for_status()
if "application/json" in response.headers.get("Content-Type", ""):
data = response.json()
results = []
for mol in data.get("molecules", []):
chembl_id = mol.get("molecule_chembl_id")
mol_structures = mol.get("molecule_structures", {})
canonical_smiles = mol_structures.get("canonical_smiles")
if chembl_id and canonical_smiles:
results.append({
"CHEMBL_ID": chembl_id,
"SMILES": canonical_smiles
})
return results
else:
print("Unexpected content type:", response.text)
except httpx.HTTPError as e:
print(f"HTTP error: {e}")
except Exception as e:
print(f"Unexpected error: {e}")
return None
# Tool 4
@mcp.tool()
async def similarity_search(molecule: str, similarity: int=80, limit: int = 10, timeout: float = 30.0, offset: int = 0):
"""
Performs a similarity search in the ChEMBL database using a SMILES string.
This function finds molecules in the ChEMBL database that are structurally
similar to the specified molecule, using the provided similarity threshold.
Parameters:
-----------
molecule : str
The SMILES, CHEMBL ID or InChIKey
similarity : int, optional
Similarity threshold as a percentage (0-100)
limit : int, optional
Maximum number of results to return (default: 10)
timeout : float, optional
Maximum time in seconds to wait for the API response (default: 30.0)
offset : int, optional
Number of results to skip for pagination (default: 0)
Returns:
--------
Optional[List[Dict[str, str]]]
A list of dictionaries containing the ChEMBL ID and canonical SMILES
for each similar molecule if successful, None otherwise.
Each dictionary has keys 'CHEMBL_ID' and 'SMILES'.
"""
encoded_molecule = quote(molecule)
url = f"https://www.ebi.ac.uk/chembl/api/data/similarity/{encoded_molecule}/{int(similarity)}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
response = await client.get(url, params={"limit": limit, "offset": offset})
response.raise_for_status()
if "application/json" in response.headers.get("Content-Type", ""):
data = response.json()
results = []
for mol in data.get("molecules", []):
chembl_id = mol.get("molecule_chembl_id")
mol_structures = mol.get("molecule_structures", {})
canonical_smiles = mol_structures.get("canonical_smiles")
if chembl_id and canonical_smiles:
results.append({
"CHEMBL_ID": chembl_id,
"SMILES": canonical_smiles
})
return results
else:
print("Unexpected content type:", response.text)
except httpx.HTTPError as e:
print(f"HTTP error: {e}")
except Exception as e:
print(f"Unexpected error: {e}")
return None
if __name__ == "__main__":
mcp.run(transport='stdio')The docstrings for tools are crucial. The LLM models skim through these documentations to determine which tool to use based on your prompts. Therefore, it is best to provide detailed docstrings that describe your tools.
4. Run the MCP server:
After satisfying with cheminformatics_mcp.py scripts, we have to do two things: run the server and add the server to the ClaudeAI configuration. To run the server, you can run this command in the Terminal. Note that your recent working directory is cheminformatics_mcp.
uv run cheminformatics_mcp.pyThen, to edit the ClaudeAI configuration, you can open your Claude Desktop, then go to Settings, then select Developer, and click on Edit Config.
All the thing you need to do is open claude_desktop_config.json. Then add this code:
{
"mcpServers": {
"cheminformatics_mcp": {
"command": "uv",
"args": [
"--directory",
"ABSOLUTE PATH TO FOLDER WHICH CONTAINS cheminformatics_mcp.py",
"run",
"cheminformatics_mcp.py"
]
}
}
}After finishing these two steps, you can re-open your Claude Desktop to check if the server is connected. If there is still an error, you can try this instead:
{
"mcpServers": {
"cheminformatics_mcp": {
"command": "ABSOLUTE PATH TO UV FILE",
"args": [
"--directory",
"ABSOLUTE PATH TO FOLDER WHICH CONTAINS cheminformatics_mcp.py",
"run",
"cheminformatics_mcp.py"
]
}
}
}5. Enjoy your MCP server:
Open Claude Desktop, ask something like: “Give me the chemical properties of CHEMBL464268”, you will have:
Previously, if you simply used ClaudeAI without connecting to the MCP server, this would be your answer:
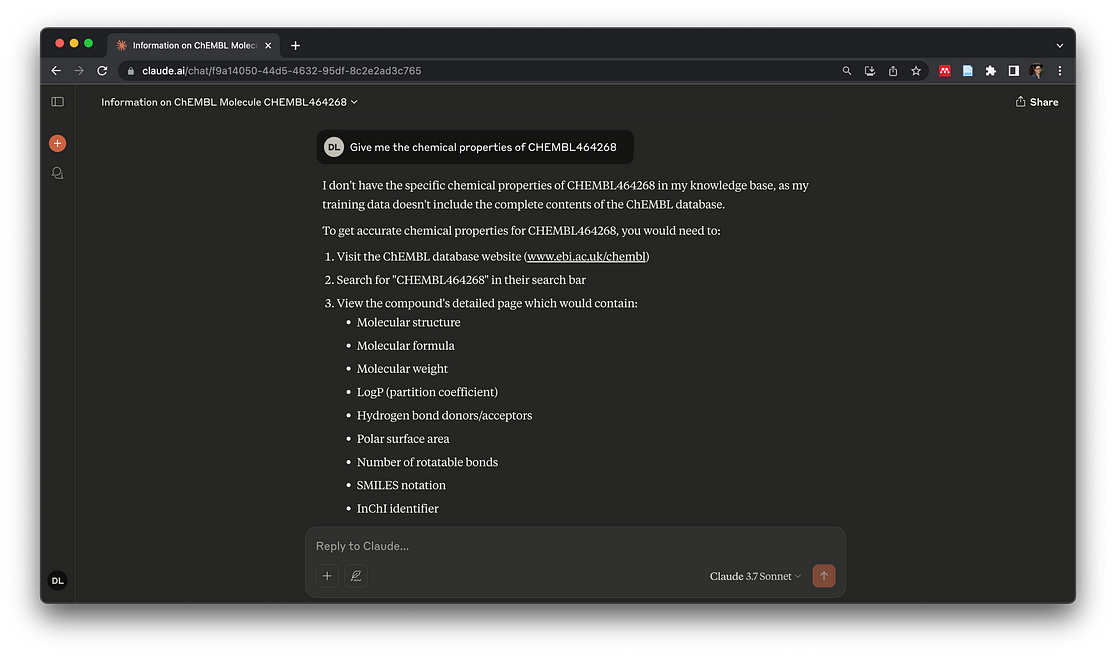
MCP IS GOOD BUT NOT PERFECT
At this point, you might be wondering about the real value of the MCP if we still need to build tools manually. The key is that AI can extend its capabilities. If we only rely on the tools for their direct outputs, we obtain results that lack context. However, with MCP:
AI can execute tasks precisely as we expect.
It can take raw outputs and transform them into deeper insights, converting basic results into meaningful understandings. In this way, MCP enables AI not only to perform tasks but also to interpret results and add value to the outcomes.
However, it is important to note that MCP is not perfect. It requires significant effort from developers to be efficient, contrary to the common media ad that suggests you can use MCP without any coding. Specifically, developers must have a good understanding of the integrated data.
In our case, this refers to the ChEMBL web service. For example, if we simply call the entire JSON file without a properly formatted function, there is a high chance that the results will exceed the conversation length limit of ClaudeAI. This means that, although the MCP server can theoretically help you complete tasks, it may not be able to run. This presents a major challenge: you need to design your tools to provide responses that are appropriately lengthy, especially not too long.
To illustrate this point, let's have a look at the first approach, which has no post-response process. If you run the following code for CHEMBL25, the result has 20035 characters.
@mcp.tool()
async def similarity_search(compound: str, similarity=80, limit: int = 10, timeout: float = 30.0, offset: int = 0):
encoded_compound = quote(compound)
url = f"https://www.ebi.ac.uk/chembl/api/data/similarity/{encoded_compound}/{int(similarity)}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
response = await client.get(url, params={"limit": limit, "offset": offset})
response.raise_for_status()
if "application/json" in response.headers.get("Content-Type", ""):
return response.json()
else:
print("Unexpected content type:", response.text)
except httpx.HTTPError as e:
print(f"HTTP error: {e}")
except Exception as e:
print(f"Unexpected error: {e}")
return NoneBut if you have a post-process, which only returns the CHEMBL_ID and SMILES of compounds, the answer now is only 457 characters, which is a significant drop compared to 20035 characters.
@mcp.tool()
async def similarity_search(smiles: str, similarity=80, limit: int = 10, timeout: float = 30.0, offset: int = 0):
encoded_compound = quote(compound)
url = f"https://www.ebi.ac.uk/chembl/api/data/similarity/{encoded_compound}/{int(similarity)}.json"
async with httpx.AsyncClient(timeout=timeout) as client:
try:
response = await client.get(url, params={"limit": limit, "offset": offset})
response.raise_for_status()
if "application/json" in response.headers.get("Content-Type", ""):
data = response.json()
# Post-response processing
results = []
for mol in data.get("molecules", []):
chembl_id = mol.get("molecule_chembl_id")
mol_structures = mol.get("molecule_structures", {})
canonical_smiles = mol_structures.get("canonical_smiles")
if chembl_id and canonical_smiles:
results.append({
"CHEMBL_ID": chembl_id,
"SMILES": canonical_smiles
})
return results
else:
print("Unexpected content type:", response.text)
except httpx.HTTPError as e:
print(f"HTTP error: {e}")
except Exception as e:
print(f"Unexpected error: {e}")
return NoneFinal thoughts
In our setup, MCP serves as a protocol for executing Python scripts using LLM. We actually access ChEMBL using Python and use ClaudeAI to execute the scripts. While we cannot deny the significant advantages of MCP in providing up-to-date data and interpreting it, developers’ efforts are still essential.
This is part one of the blog; in parts two and three, we will continue with the PubChem and PDB databases.
Comments
Post a Comment